Research Group: Dynamic Metabolons
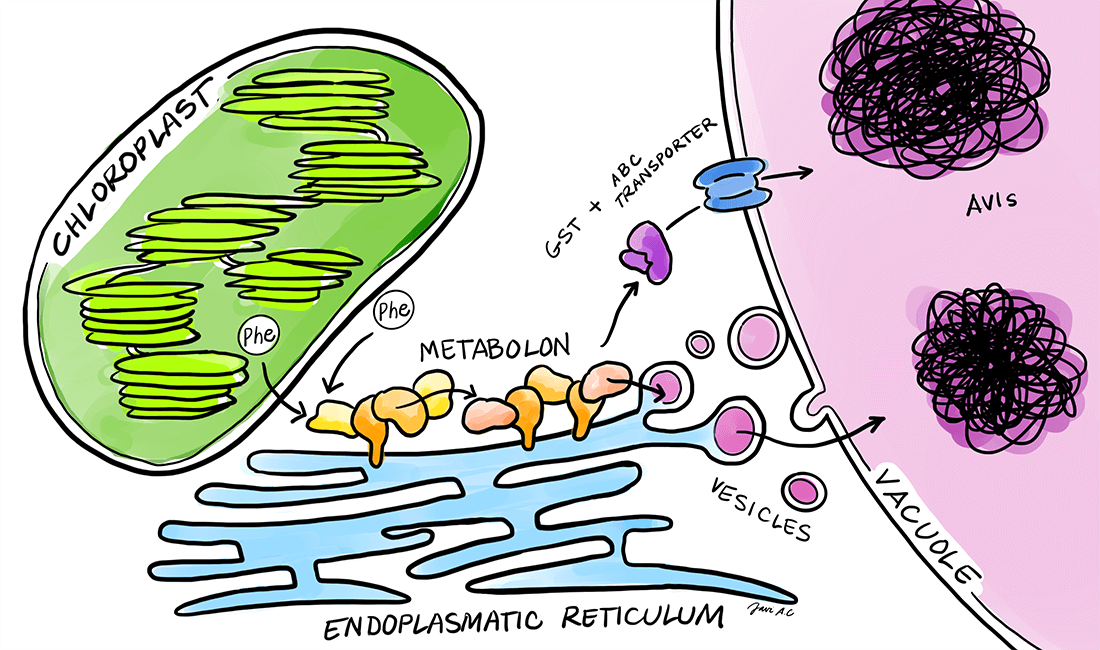
Plants produce an immense number of bioactive compounds with an immense potential for applications as pharmaceuticals, colorants, flavors etc. The chemical diversity of phytochemicals is driven by modular assembly of different enzyme classes such as oxidoreductases, cytochromes P450 and transferases. Assembly of enzymes in efficient complexes, termed metabolons, facilitates direct transfer of substrates and intermediates between sequential enzymes, which results in increased flux devoid of concomitant release of toxic or labile intermediates. We are interested in understanding how plants assemble metabolons on-demand in response to environmental challenges. As biological systems we are focusing on cyanogenic glucoside biosynthesis, dhurrin, in Sorghum bicolor and flavonoid biosynthesis in Petunia axillaris and Petunia inflata.
- Discovery of new proteins involved in regulation of metabolon assembly
-
We are interested in general mechanisms regulating the assembly of metabolons such as scaffolding proteins, protein-protein interactions and the local membrane environment. In order to identify new regulatory proteins, we take an untargeted approach based on proximity labeling combined with mass spectrometry-based proteomics.
- Dynamic assembly and structural organization
-
We are uncovering the dynamics of metabolon assembly at the single molecule level. The diffusion of individual enzymes and entire metabolons provides kinetic insight on the assembly and disassembly. In the process, we are developing new platforms for studying protein-protein interactions, membrane sorting and binding of soluble enzymes. We are also studying the structural organization of metabolons and their different configurations by Cryo-EM, Crystallography and Neutron scattering techniques.
- Functional consequences
-
We apply our findings to study the functional consequences of enzyme engineering on substrate channeling using in vivo and in vitro systems. Reconstitution of entire biosynthetic pathways in yeast allows for analysis of new regulatory proteins and their role on substrate channeling using mass spectrometry-based metabolomics. As part of this research topic we are optimizing biomimetic membrane systems for studying multienzyme complexes.
For students wishing to work within these topics. M.Sc. and B.Sc. projects are currently available. Please contact tola@plen.ku.dk
2022-2024 FLAVOBIO, Eurostars Programme (Co-PI: Tomas Laursen)
2019-2024 The language of plant metabolic highways, Emerging Investigator Novo Nordisk Foundation (PI: Tomas Laursen)
2018-2022 Directing plant metabolism towards formation of high value bioactive products, Sapere Aude Starting Grant, Independent Research Fund Denmark (PI: Tomas Laursen)
- Biased cytochrome P450-mediated metabolism via small-molecule ligands binding P450 oxidoreductase
Jensen, S.B., Thodberg, S., Parween, S., Moses, M.E., Hansen, C.C., Thomsen, J., Sletfjerding, M.B., Knudsen, C., Del Giudice, R., Lund, P.M., Castano, P.R., Bustamante, Y.G., Velaquez, M.N.R., Jørgensen, F.S., Pandey, A.V., Laursen, T., Møller, B.L., Hatzakis, N.S.
(2021) Nature Communications 12:2260
Link: https://www.nature.com/articles/s41467-021-22562-w.pdf
- Membrane anchoring facilitates colocalization of enzymes in plant cytochrome P450 redox systems
Laursen, T., Lam, HYM., Sørensen, K.K., Tian, P., Hansen, C.C., Groves, J.T., Jensen, K.J., Christensen, S.M
(2021) Communications Biology 4:1057
Link: https://www.nature.com/articles/s42003-021-02604-1.pdf
- Stabilization of dhurrin biosynthetic enzymes from Sorghum bicolor using a natural deep eutectic solvent
Knudsen, C., Bavishi, K., Viborg, K.M., Drew, D.P., Simonsen, H.T., Motawia, M.S., Møller, B.L., Laursen, T.
(2020) Phytochemistry 170:112214
Link: https://www.sciencedirect.com/science/article/pii/S0031942219308040?via%3Dihub
- Molecular snapshots of dynamic membrane-bound metabolons
Bassard, J.E., Laursen, T.*
(2019) Methods in Enzymology 617, 1-27
Link: https://www.sciencedirect.com/science/article/pii/S007668791830497X
- Dynamic metabolic solutions to the sessile life style of plants
Knudsen, C., Gallage, N.J., Hansen, C.C., Møller, B.L., Laursen, T.*
(2018) Natural Product Reports 35()11, 1140-1155
Link: https://pubs.rsc.org/en/content/articlepdf/2018/np/c8np00037a
- Direct observation of multiple conformational states in Cytochrome P450 oxidoreductase and their modulation by membrane environment and ionic strength
Bavishi, K., Li, D., Eiersholt, S., Hooley, E., Petersen, T., Møller, B.L., Hatzakis, N.S.*, Laursen, T.*
(2018) Scientific Reports, 8(1), 6817
Link: https://www.nature.com/articles/s41598-018-24922-x.pdf
- Controlling Styrene Maleic Acid Lipid Particles through RAFT
Smith A.A.A., Autzen H.E., Laursen T., Wu V., Yen M., Hall A., Hansen SD., Cheng Y., Xu T.
(2017) Biomacromolecules, 13;18(11), 3706-3713
Link: https://pubs.acs.org/doi/pdf/10.1021/acs.biomac.7b01136?rand=m79vo748
- Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum
Laursen, T., Borch, J., Knudsen, C., Bavishi, K., Torta, F., Martens, H.J., Silvestro, D., Hatzakis, N.S., Wenk, M.R., Dafforn, T.R., Olsen, C.E., Motawia, M.S., Hamberger, B., Møller, B.L., Bassard, J-E.
(2016) Science 354(6314), 890-893
Link: https://science.sciencemag.org/content/354/6314/890/tab-pdf
- Single molecule activity measurements of cytochrome P450 oxidoreductase reveal the existence of two discrete functional states
Laursen, T.*, Singha, A.*, Rantzau, N., Tutkus, M., Borch, J., Hedegard, P., Stamou, D., Møller, B.L., Hatzakis, N.S.
(2014) ACS Chemical Biology 9(3), 630-634
Link: https://pubs.acs.org/doi/pdf/10.1021/cb400708v?rand=a7xkmkq5
Our research group takes an interdisciplinary basic science approach to elucidate fundamental mechanisms governing the plasticity of the biosynthesis of phytochemicals, which is well positioned between multiple groups at the Section for Plant biochemistry working on pathway discovery and heterologous production, including:
- Biochemical engineering (Sotirios Kampranis)
- Alkaloids and Chemical Ecology (Fernando Geu Flores)
- Synthetic Biology (Birger Lindberg Møller)
- Molecular Evolution of Specialized Metabolism (Søren Bak)
Our basic science approach benefits from collaboration with additional internal groups with expertise in plant physiology and metabolomics, including:
- Plant Ecophysiology (Elisabeth Heather Neilson)
- Plant Metabolic plasticity (Nanna Bjarnholt)
We are actively collaborating with world leading external experts, which provide access to state-of-the-art technologies and knowledge, including:
- Petunia genetics (Ronald Koes and Francesca Quattrocchio, University of Amsterdam, NL)
- Cryo Electron Microscopy (Bjørn Panyella Pedersen, Aarhus University, DK)
- Native Mass Spectrometry (Jonathan T. Hopper and Carol Robinson, Oxford University, UK)
- Neutron Reflectometry (Marité Cardenas, Malmö University, SE)
- SMALP technology (Timothy Dafforn, University of Birmingham, UK)
- Single Molecule Microscopy (Jay T. Groves, UC-Berkeley, USA and Nikos Hatzakis, University of Copenhagen, DK)
- Computational modeling (Flemming Steen Jørgensen, University of Copenhagen, DK)
- Ligand Fishing and NMR (Dan Stærk, University of Copenhagen, DK)
We have initiated several collaborations with industrial partners on heterologous production of flavonoids
- Anthocyanin production in yeast (Lantana Bio, FR)
- Flavonol production in yeast (Biosyntia, DK)
Group members
| Name | Title | Phone | |
|---|---|---|---|
| Tomas Laursen | Associate Professor | +4535333329 |
Master students
- Clara Ross Zaulich
- Kees Buhrman

